Scaling the Neo4J Write Path – Part 1
- Software Engineering
Neo4J is one of the most popular graph databases in the world. If you explore the space you very quickly discover that many different graph databases exist, spanning from basic single machine triplestores like Apache Jena TDB to horizontally scalable graph databases such as Janus or DGraph and far too many others to list here, varying not only in their scalability but in their approaches to linked data modelling.
All of these options are useful in their own way and have their own selling points. If you explore the community you will quickly find that Neo4J in particular has an interesting reputation. It has been around for a long time, relative to the space and as such it has by far the richest ecosystem around it.
Unlike most projects, our use case needed users to be able to issue arbitrary queries against the graph data, with a schema that dynamically adapted to new information. We also needed users to be able to perform complex statistical analyses, as well as being able to explore the results themselves without having to involve the dev teams. While we considered a large number of systems and created proof of concepts for several, Neo4J was the natural target for a wide range of reasons.
One G-Research’s guiding principles is that its people are empowered to solve hard problems. While I won’t go into our reasoning in any detail, it was clear that Neo4J was the best choice, if it could be made to handle the volumes of writes we needed. This had seemingly not been previously attempted. When I presented the options and trade-offs, I was empowered to embark on this journey, working directly with Neo4J Inc to push the limits and scale it further than it had ever been scaled before.
Sharding
Neo4J has one big limitation which in my experience is often the main reason it gets discounted: Neo4J is vertically scalable!
The database uses “Graph Native Storage” which is optimised for extremely fast queries and is able to deliver on that promise better than its competition because it simply doesn’t allow you to shard your data amongst nodes. It is very efficient and you can easily have a graph database with billions of nodes on a single machine. You can also scale the read performance by having many read-only followers. But still, every node must have 100% of the data.
These days it is possible to get machines sporting terabytes of memory so this is not an insurmountable problem. For our POC I obtained one such machine. So while I might be nervous about being able to store everything long-term as our multi-terabyte dataset grows it was ‘fine’ for the POC.
You could have multiple databases with a fraction of the data in each one. But should you need to query cross database things get messy.
Fabric
Neo4J 4.0 introduced Fabric to get around this. Fabric provides exactly this: a way to query data that is split across databases. This can be used to combine data from logically separate graphs or as a means to shard a large graph into sub-graphs, allowing horizontal scalability. This means as a data publisher you must partition your data and as a user the fact that data is distributed cross database leaks into your queries as you specifically query multiple databases and union the results. This is a bit messy and also restricted to Neo4J’s enterprise edition. Keeping in mind our goal to have users access this themselves, we would rather avoid this complexity.
The Single Writer Limit
Having to host all data on one big machine is okay from a read perspective. This is not dissimilar to most SQL databases that run on huge machines and, like Neo4J, provide good performance and ACID properties.
The big issue that arises is the write-path. Neo4J can have many read-only copies of the data but even if you use the causal clustering there is only ever a single leader which can apply writes. For many use-cases this is fine. Most people probably add links and properties a few at a time over the course of a day. If you are dealing with millions per second you might have some issues. Our dataset comes in once a day and is approximately 500GB (compressed) of triples. While for the initial use-case we could filter this down and only apply changes, we expect to be dealing with far larger volumes of data and we will need to add a similar large initial load for subsequent datasets included in the graph. This means we needed to know that Neo4J will scale sufficiently before we commit to it.
To help us out we had two excellent Neo4J consultants assisting us on this journey.
How fast can we load this data?
Ideally, we would load the data as quickly as possible. If we were using something like JanusGraph or Neo4J Fabric we could simply scale out our cluster until we hit the performance we were looking for. In Neo4J, without Fabric, we don’t have that luxury so for the POC we would settle for being able to keep up. That just means that we have to be able to load 24 hours of data within 24 hours. Clearly, the faster the better but that is our goal.
The neosemantics tool
Neo4J provides a tool called neosemantics, or n10s, which is specifically designed for loading triples into Neo4J. I don’t actually know how long this would take to load the data with its default settings but suffice to say I left it running over a weekend and it hadn’t finished on Monday.
Fortunately, there are various options for tuning so we can tweak the commit size and cache size to minimise db accesses. With sufficient tuning to take advantage of our huge test machine we were able to get this to load in three days. Sadly that means it is not viable.
Analysing the process (and with some help from some performance experts from Neo4J) we realised there were two issues with this approach:
- Neo4J is transactional and writing 500G in transactions, even large ones, is going to take a long time and cripple read performance in the process
- The library is doing three tasks; reading the triples, parsing them and finally loading them
This led to two realisations:
- We need to find a way to reduce the impact of transactions
- Reading and parsing the triples are embarrassingly parallel problems but we are restricted to a single node. By letting Spark do the hard work of pre-processing the data, all we have to do is load it
Spark app
We first tried simply pre-processing the data on Spark which took around ten minutes. However, writing the result to Neo4J from Spark was not finished after 24 hours so we abandoned this approach.
Neo4J now provide an official Spark connector. When we originally performed this test we used the now deprecated CAPS project. CAPS was inherently limited both on read and write side. While the test has not been re-run using the new connector, as pointed out in the FAQs fundamentally the spark connector still writes in transactional batches and is thus exactly as limited as the prior attempt using n10s.
Following the parallelism best practices if we optimally divided our data into non-overlapping segments we could at most have parallelism equal to the number of cores on the machine. With over 500G of data on a machine with 128 vcores we are still looking at 4G of heavily compressed data per partition and while the write was hammering the database, read performance (as well as non-batch writes) would be severely impacted.
There was also a project named Morpheus which aimed to store Neo4J data on HDFS but this is sadly defunct. GraphFrames is another interesting option along similar lines which could allow us to keep our golden data on HDFS. We considered having users produce views which could be loaded into dedicated Neo4J instances for OLTP workloads. However, this adds to the complexity and means we don’t have all the data available in Neo4J.
Spark app + batch loader
Going back to our key takeaways from the n10s experiment, we are now pre-processing our data on Spark so we’ve removed most of the work from the loader. The remaining bottleneck is the single writer and its transactional nature.
In SQL databases such as MS SQL Server, you can drastically speed up loading huge quantities of data by turning off the WAL and replication then re-snapshotting once you are done. Looking into this for Neo4J it is not quite so simple. The disk structure is cleverly optimised to help with traversal performance but by doing so we can’t simply cheat by using unlogged batches or doing the equivalent of shoving an SSTable on disk and telling Neo4J about it later. This sort of approach can be used to great effect in systems such as HBase to bypass the slow write path entirely.
The closest equivalent in Neo4J is using the neo4j-admin import. Whilst this does not allow bulk loading to an active database it promises the fastest way to write large quantities of data to Neo4J. What it really does is essentially what I described above except for the entire database not just the new data. It makes a totally new set of database files at which we can then point Neo4J and it will start up happily. Doing this means we are no longer restricted by that pesky transaction manager. We don’t even have to do this on the same node, we can do it anywhere and just put the resulting data in the right place later.
It expects the data in a CSV format split such that each type of node gets a file with all the possible properties for this node and each relationship gets a similar file. In our case the structure of the data is actually variable. The number of nodes and properties is completely unknown but fortunately through sufficient work in Spark it is possible to generate this large batch of CSVs.
After pre-processing, we had 300G of CSV files. On our beefy machine the admin-loader was able to generate the new db in just over a day (27 hours). This is close, with a sufficiently beefier machine using NVMe SSDs this would probably be fast enough for now. But that is uncomfortably close to our limits and this is only our first dataset in this system.
Spark App + Internal BatchImporter Custom Loader
Having been so close with the prior attempt, we profiled the loader and it turned out most of the time is actually spent reading the data and processing the CSVs. The CSVs are strings which is not only inefficient storage but means that any binary types like integers and floats which make up a lot of the dataset need converting.
Our performance specialist friends at Neo4J suggested trying out the BatchImporter which is part of the “internal” Java package. This is actually exactly what the neo4j-admin import tool uses under the bonnet as you can see in the code. If you follow the code through, the ParallelBatchImporter is where the magic happens. Looking at that class the comment says it all:
“…tries to exercise as much of the available resources to gain performance. Or rather ensure that the slowest resource (usually I/O) is fully saturated and that enough work is being performed to keep that slowest resource saturated all the time.”
The biggest caveat of this API is you can touch each node and relation exactly once!
The other concern an astute reader might have noticed is that this is using an internal API. We discussed this with our Neo4J contacts and they assured us it should be reasonably stable.
The internal package is actually a fairly recent repackaging. This used to all be in the unsafe package because the key aspect is that you are not using the actual Neo4J API, you are dealing directly with the backing stores. It assumes if you’re using the API, you know what you’re doing; be warned it is very easy to end up with an invalid file!
So we know that we are IO-bound. We also know we waste a lot of CPU time on parsing. The logical next step is to make our own version of the CSV importer but with those issues minimised.
- The loader is a Java API so we want Java types. We therefore create a custom Hadoop Writable which allows us to collect all the data we need for a node into a single object, being careful to preserve the property types
- A single machine has to read all the data so we want to compress it well but optimise for read speed. LZ4 is a great pick for this but is slightly awkward as the LZ4 files written by Hadoop are unframed and as such cannot be read by most libraries. We also want to make sure we use a splittable file format which supports block compression so our choice was the Hadoop SequenceFile format.
To cut a long story short this works really well. The native storage takes us down from that initial 500G input to a mere 50G of LZ4 data. Inserting this data using our custom loader takes a mere 40 minutes on my dev machine. Running it on the chunkier machine takes it down to only 20 minutes!
Could we do better?
At first glance this is looking good, we’re reading quite fast right from the start and CPU is looking pretty saturated.
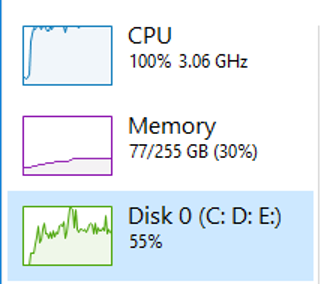
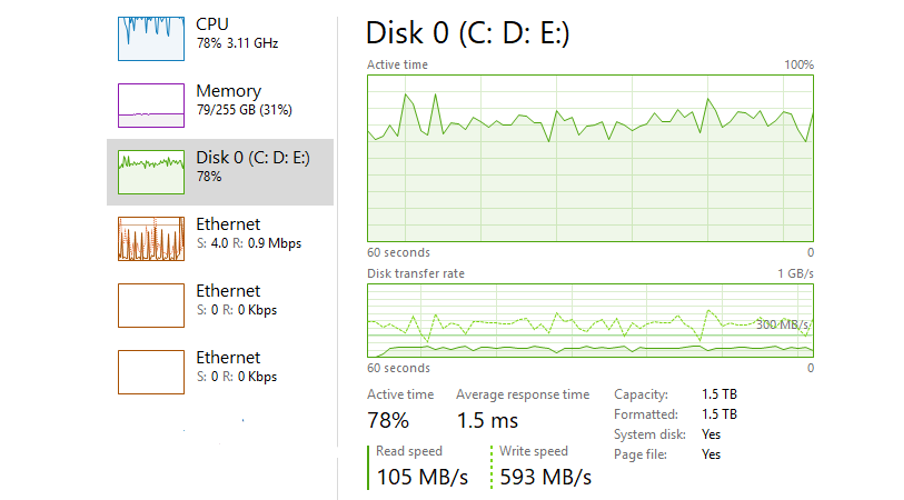
After we let it settle into a comfortable rhythm it is evident that we aren’t quite maxing out my machine with this method. I’ve given the Java process 100G of RAM but it is only using ~75GB, my disk is much better utilised but not our limit. It is clear that CPU is still our bottleneck here.
Using Java Flight Recorder we can dig into where all of this time is going and this is what we see:
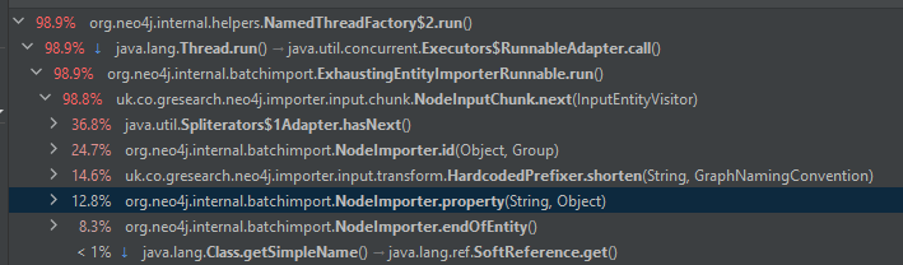
My first takeaway from this is that we’ve drastically reduced most of the overheads. Almost half of our time is now spent just creating data (45.8 from just those NodeImporter methods). A bit more than a third is also spent reading which means we have at least made it so the majority of the time is not taken reading and parsing CSVs.
It is clear this is far from optimal. I will cover potential improvements in a future post but we’ve already far exceeded the traditional single machine limits.
Technical Details
This section aims to cover some of the more interesting details of the implementation.
The Custom NodeWritable Output Format
One of the biggest wins in this process is the ability to avoid the need to parse strings into their real types as part of the importer. While we are still paying some overheads, being able to send the data in a JVM friendly format is very powerful. This approach can be extended to have Spark or Hadoop write out any format of data you like.
In our particular implementation the biggest challenge was in allowing a heterogeneous property map to make it through while preserving the type information and avoiding needing to convert via some intermediary format like a string. Recall that you can only visit a node exactly once! This means you need all of your properties together and on the import side you need to be able to add properties one by one with the appropriate types.
The Heterogeneous Map of Homogeneous Lists
Java is a mostly strongly, statically typed language. As such it enforces that any container type like a list or map must have objects of only a single type. We want to preserve the types of the properties.
While we could store our properties as Map<String, List<Object>> this would lose that valuable type information and would allow a list mixing integers and strings which would fail on import when it is cast to a concrete list of a specific type. So instead we use Java’s wild card generics. This allows us to say Map<String, List<?>>. The ‘?’ here indicates to Java that the list will be of a homogenous type. We don’t know which one, but it will be consistent within itself. The beauty of this is that Object opens the door to heterogeneous lists which are invalid and would cause runtime exceptions when we try to cast them back to their proper types. Whereas by using the wildcard, Java ensures that all of our lists are valid and we have been able to use a single type to express this.
Java has type erasure which means that at runtime all this type information is sadly lost. Despite this the wildcard allows us to stay safe. Because the type could be anything, when we come to write these property lists we have to check the type of the first item, then cast accordingly. We could resolve this by using a wrapper to record the type of the list but simply checking the first item is fine as we know all items must be of the same type.
The Hadoop Writable interface does not support writing complex types such as lists or arrays. We can, however, take a cue from how strings are serialised and use the same trick for lists as well as transmitting the types themselves. After all a string is essentially just a list of characters. To that end we first write the number of properties we are about to serialise, then for each one we write the key, the type using a helpful enum, then the number of entries for that property, and finally the actual values. On the read side the process then becomes:
- Read number of properties
- Initialise a map of the perfect size
- Until we hit the expected number of properties:
- Read the property key (
String) - Read the property type (
Enum) - Use the parser for the detected type to deserialise the list
- Read the number of values expected
- Initialise a list of the perfect capacity
- Read the property key (
- Read values of the appropriate type until we hit the expected number
Due to erasure we still are left with a Map<String, List<?>> but that’s okay because the Neo4J property interface actually just accepts Object.
public class NodeWritable implements Writable {
private String nodeId;
private Map<String, List<?>> properties;
public NodeWritable() {
this(null);
}
public NodeWritable(String nodeId) {
this(nodeId, new HashMap<>());
}
public NodeWritable(String nodeId, Map<String, List<?>> properties) {
this.nodeId = nodeId;
this.properties = properties;
}
public String getNodeId() {
return nodeId;
}
public void setNodeId(String nodeId) {
this.nodeId = nodeId;
}
public Map<String, List<?>> getProperties() {
return properties;
}
public void setProperties(Map<String, List<?>> properties) {
this.properties = properties;
}
@Override
public String toString() {
return "Node{" +
"nodeId='" + nodeId + ''' +
", properties=" + properties +
'}';
}
@Override
public void write(DataOutput out) throws IOException {
writeNodeId(out);
writeProperties(out);
}
private void writeProperties(DataOutput out) throws IOException {
WritableUtils.writeVInt(out, properties.size());
for (Map.Entry<String, List<?>> entry : properties.entrySet()) {
WritableUtils.writeString(out, entry.getKey());
writeObjectList(out, entry.getValue());
}
}
private void writeObjectList(DataOutput out, List<?> list) throws IOException {
Object value = list.get(0);
if (value instanceof String) {
WritableUtils.writeEnum(out, PropertyTypes.STRING);
PropertyTypes.STRING.writeList(out, list);
} else if (value instanceof Byte) {
WritableUtils.writeEnum(out, PropertyTypes.BYTE);
PropertyTypes.BYTE.writeList(out, list);
} else if (value instanceof Long) {
WritableUtils.writeEnum(out, PropertyTypes.LONG);
PropertyTypes.LONG.writeList(out, list);
} else if (value instanceof Integer) {
WritableUtils.writeEnum(out, PropertyTypes.INT);
PropertyTypes.INT.writeList(out, list);
} else if (value instanceof Double) {
WritableUtils.writeEnum(out, PropertyTypes.DOUBLE);
PropertyTypes.DOUBLE.writeList(out, list);
} else if (value instanceof Float) {
WritableUtils.writeEnum(out, PropertyTypes.FLOAT);
PropertyTypes.FLOAT.writeList(out, list);
} else if (value instanceof Boolean) {
WritableUtils.writeEnum(out, PropertyTypes.BOOL);
PropertyTypes.BOOL.writeList(out, list);
} else if (value instanceof Character) {
WritableUtils.writeEnum(out, PropertyTypes.CHAR);
PropertyTypes.CHAR.writeList(out, list);
} else if (value instanceof Short) {
WritableUtils.writeEnum(out, PropertyTypes.SHORT);
PropertyTypes.SHORT.writeList(out, list);
} else {
throw new RuntimeException("Property Map Entry with un-serializable type, Sample entry: " + value.toString());
}
}
public void addProperty(String key, List<?> list) {
if (properties.containsKey(key)) {
throw new RuntimeException("Each property should only be set once");
}
properties.put(key, list);
}
private Map<String, List<?>> readPropertyMap(DataInput in) throws IOException {
int properties = WritableUtils.readVInt(in);
Map<String, List<?>> propertyMap = new HashMap<>(properties, 1.0f);
for (int property = 0; property < properties; property++) {
String key = WritableUtils.readString(in);
PropertyTypes type = WritableUtils.readEnum(in, PropertyTypes.class);
List<?> entryVal = type.readList(in);
propertyMap.put(key, entryVal);
}
return propertyMap;
}
private void writeNodeId(DataOutput out) throws IOException {
WritableUtils.writeString(out, nodeId);
}
@Override
public void readFields(DataInput in) throws IOException {
nodeId = readNodeId(in);
properties = readPropertyMap(in);
}
private String readNodeId(DataInput in) throws IOException {
return WritableUtils.readString(in);
}
}
public enum PropertyTypes {
STRING {
@Override
void writeList(DataOutput out, List<?> list) throws IOException {
writePropertyListSize(out, list);
for (String item : (List<String>) list) {
WritableUtils.writeString(out, item);
}
}
@Override
List<String> readList(DataInput in) throws IOException {
int entries = readPropertyListSize(in);
List<String> list = new ArrayList<>(entries);
for (int entry = 0; entry < entries; entry++) {
list.add(entry, WritableUtils.readString(in));
}
return list;
}
},
BYTE {
@Override
void writeList(DataOutput out, List<?> list) throws IOException {
writePropertyListSize(out, list);
for (Byte item : (List<Byte>) list) {
out.writeByte(item);
}
}
@Override
List<Byte> readList(DataInput in) throws IOException {
int entries = readPropertyListSize(in);
List<Byte> list = new ArrayList<>(entries);
for (int entry = 0; entry < entries; entry++) {
list.add(entry, in.readByte());
}
return list;
}
},
// Other types redacted for simplicity
SHORT {
@Override
void writeList(DataOutput out, List<?> list) throws IOException {
writePropertyListSize(out, list);
for (Short item : (List<Short>) list) {
out.writeShort(item);
}
}
@Override
List<Short> readList(DataInput in) throws IOException {
int entries = readPropertyListSize(in);
List<Short> list = new ArrayList<>(entries);
for (int entry = 0; entry < entries; entry++) {
list.add(entry, in.readShort());
}
return list;
}
};
void writePropertyListSize(DataOutput out, List<?> list) throws IOException {
int entries = list.size();
WritableUtils.writeVInt(out, entries);
}
int readPropertyListSize(DataInput in) throws IOException {
return WritableUtils.readVInt(in);
}
abstract void writeList(DataOutput out, List<?> list) throws IOException;
abstract List<?> readList(DataInput in) throws IOException;
}
There are a number of issues with this implementation, not least that EnumWritables are surprisingly inefficient. However such improvements will be discussed in a later article for those interested.
Reading LZ4 Data
One snag when creating this app was the serialisation format. A brief search will quickly show that, in terms of the standard Spark compression options, LZ4 is a clear winner for our use-case. It is splittable, has decent compression and is fast to read. However Hadoop’s usage of LZ4 is slightly complex to utilise.
LZ4 has a framed compression format or a block compression format. Most tools for reading LZ4 files such as unlz4 (which is available on most Linux systems) cannot read LZ4 files produced by the Hadoop Lz4Codec.
The cause of this complexity is this open Hadoop ticket: https://issues.apache.org/jira/browse/HADOOP-12990
Essentially, Hadoop hard-codes some context and thus does not provide the correct frame header you get from a normal LZ4 file. For example, if I take a Hadoop log which I’ve already decompressed from LZ4 and recompress it with the same 256K block size as Hadoop uses by default:
lz4 -B5 /mnt/e/application2.log
Examining the header, it looks like this:

This is the frame header. But looking at the same file compressed using LZ4 from Hadoop we see:

Note that Hadoop writes the block format directly.
As such you either need to use the LZ4BlockInputStream from lz4-java or simply use the Hadoop LZ4 Codec directly.
/**
* Decompress an LZ4 Block compressed stream using lz4-java
*/
public InputStream transformLz4Java(InputStream inputStream) throws IOException {
return new LZ4BlockInputStream(inputStream, LZ4Factory.fastestInstance().fastDecompressor());
}
// -------------------------------------------------------------------- //
private static final Configuration HDFS_CONF = new Configuration(true);
static {
System.load("C:/ProgramData/Hadoop/bin/hadoop.dll");
System.load("C:/ProgramData/Hadoop/bin/hdfs.dll");
System.setProperty("HADOOP_COMMON_LIB_NATIVE_DIR", "C:/ProgramData/Hadoop/lib");
System.setProperty("HADOOP_OPTS", "-Djava.library.path=C:/ProgramData/Hadoop/lib");
}
/**
* Decompress an LZ4 Block compressed stream using Hadoop native lib
*/
public CompressionInputStream transformHadoopLib(InputStream inputStream) throws IOException {
Lz4Codec lz4Codec = new Lz4Codec();
lz4Codec.setConf(HDFS_CONF);
Decompressor d = lz4Codec.createDecompressor();
return lz4Codec.createInputStream(inputStream, d);
}
The Batch Importer
Creating your own version of the CSVImporter is not hard. You can essentially copy it, only switching out the InputIterables. As you can see from the code below, there are plenty of dials to tune and details specific to our use case have been redacted.
public static void main(String[] args) throws Throwable {
Groups groups = new Groups();
groups.getOrCreate("MyGroup");
InputIterable nodes = () -> new FileBasedChunkInputIterator<>("E:/nodeData", groups, NodeFileParser::new, NodeInputChunk::new);
Supplier<ChunkFileParser<Quad>> lz4AwareQuadFileReader = () -> new QuadFileParser(LZ4DecompressingTransformer::new);
InputIterable rels = () -> new FileBasedChunkInputIterator<>("E:/relData", groups, lz4AwareQuadFileReader, RelationshipInputChunk::new);
Config dbConfig = Config.defaults();
File badFile = new File("F:/badLines.txt");
badFile.renameTo(new File(badFile.toString().concat("." + System.currentTimeMillis())));
File storeDir = new File("C:/output/data.db");
FileUtils.deleteDirectory(storeDir);
storeDir.mkdirs();
try (FileOutputStream badLines = new FileOutputStream(badFile);
BadCollector badCollector = new BadCollector(badLines, BadCollector.UNLIMITED_TOLERANCE, 1)) {
BatchImporter importer = BatchImporterFactory.withHighestPriority().instantiate(
DatabaseLayout.ofFlat(storeDir),
new DefaultFileSystemAbstraction(),
null,
PageCacheTracer.NULL,
new Configuration.Overridden(Configuration.DEFAULT) {
@Override
public int batchSize() {
return 1000000;
}
@Override
public boolean highIO() {
return true;
}
},
new SimpleLogService(NullLogProvider.getInstance(), NullLogProvider.getInstance()),
ExecutionMonitors.defaultVisible(),
AdditionalInitialIds.EMPTY,
dbConfig,
RecordFormatSelector.selectForConfig(dbConfig, NullLogProvider.getInstance()),
new PrintingImportLogicMonitor(System.out, System.err),
JobSchedulerFactory.createInitialisedScheduler(),
badCollector,
TransactionLogInitializer.getLogFilesInitializer(),
EmptyMemoryTracker.INSTANCE);
long start = System.currentTimeMillis();
importer.doImport(Input.input(
nodes,
rels,
IdType.STRING,
// Be sure to provide reasonable estimations
Input.knownEstimates(0, 0, 0, 0, 0, 0, 0),
groups));
System.out.println("Done after " + ((System.currentTimeMillis() - start) / 1000) / 60 + "m");
}
}
One significant optimisation here is that we avoid re-creating new input chunks for each file by switching out the underlying streams when one runs out. This is what the FileBasedChunkInputIterator does:
public class FileBasedChunkInputIterator<T> implements InputIterator {
private static final Logger LOG = LoggerFactory.getLogger(FileBasedChunkInputIterator.class);
private final Iterator<Path> chunksStream;
private final Groups groups;
private final Supplier<ChunkFileParser<T>> parserSupplier;
private final ChunkFactory<T> chunkFactory;
public FileBasedChunkInputIterator(String filesFolder, Groups groups, Supplier<ChunkFileParser<T>> parserSupplier, ChunkFactory<T> chunkFactory, Predicate<Path> fileFilter) {
this.groups = groups;
this.parserSupplier = parserSupplier;
this.chunkFactory = chunkFactory;
try {
chunksStream = Files.list(Paths.get(filesFolder))
.filter(fileFilter)
.collect(Collectors.toList()).iterator();
} catch (IOException e) {
throw new RuntimeException("Failed to read files", e);
}
}
public FileBasedChunkInputIterator(String filesFolder, Groups groups, Supplier<ChunkFileParser<T>> parserSupplier, ChunkFactory<T> chunkFactory) {
this(filesFolder, groups, parserSupplier, chunkFactory, file -> true);
}
@Override
public InputChunk newChunk() {
LOG.info("New chunk created");
Stream<T> nextChunkStream = nextChunkStream();
if (nextChunkStream != null) {
return chunkFactory.createChunk(nextChunkStream, groups);
}
return chunkFactory.createChunk(Stream.empty(), groups);
}
@Override
public boolean next(InputChunk chunk) throws IOException {
//noinspection unchecked Internally this is only called using a chunk from this instance and thus this cast is safe.
RefillableInputChunk<T> inputChunk = (RefillableInputChunk<T>) chunk;
if (!inputChunk.hasNext()) {
Stream<T> newChunk = nextChunkStream();
if (newChunk != null) {
inputChunk.setStream(newChunk);
return true;
}
return false;
} else {
return true;
}
}
private Stream<T> nextChunkStream() {
if (chunksStream.hasNext()) {
synchronized (this) {
if (chunksStream.hasNext()) {
Path next = chunksStream.next();
LOG.info(next.toString());
return readFile(next);
}
}
}
return null;
}
private Stream<T> readFile(Path path) {
try {
return parserSupplier.get().readFile(path.toFile());
} catch (IOException e) {
throw new RuntimeException("Failed to parse file: " + path.toString(), e);
}
}
@Override
public void close() throws IOException {
// Not relevant
}
}
This allows us to use a pool of input chunks and just share the input streams between the chunks. The remaining magic is in the input chunks. These are passed visitors. The chunk must determine if the visitor is relevant to it and if so take action, in this case by importing a new node entity.
public class NodeInputChunk implements RefillableInputChunk<NodeWritable> {
private static final String URI_NEO_PROPERTY = "uri";
private static final String LABEL_PROPERTY = "<" + RDF.TYPE + ">";
public static final String RESOURCE_LABEL = "Resource";
// You would want to move this to the Spark phase but leaving it here allows simpler iterations
private final Prefixer prefixer = new HardcodedPrefixer(DEFAULT_NEW_PREFIX_HANDLER);
private final Group group;
private Iterator<NodeWritable> nodeWritableStream;
public NodeInputChunk(Stream<NodeWritable> nodeWritableStream, Groups groups) {
this.nodeWritableStream = nodeWritableStream == null ? Collections.emptyIterator() : nodeWritableStream.iterator();
this.group = groups.getOrCreate("MyGroup");
}
@Override
public boolean hasNext() {
return nodeWritableStream.hasNext();
}
@Override
public boolean next(InputEntityVisitor visitor) throws IOException {
if (nodeWritableStream.hasNext() && visitor.getClass().getSimpleName().equals("NodeImporter")) {
NodeWritable node = nodeWritableStream.next();
Map<String, List<?>> properties = node.getProperties();
visitor.id(node.getNodeId(), group);
visitor.property(URI_NEO_PROPERTY, node.getNodeId());
for (Map.Entry<String, List<?>> entry : properties.entrySet()) {
List<?> values = entry.getValue();
int size = values.size();
String key = entry.getKey();
if (size == 1) {
if (key.equals(LABEL_PROPERTY)) {
visitor.labels(new String[]{prefixer.shorten((String) values.get(0), GraphNamingConvention.LABEL), RESOURCE_LABEL});
} else {
visitor.property(prefixer.shorten(key, GraphNamingConvention.PROPERTY_KEY), values.get(0));
}
} else {
if (key.equals(LABEL_PROPERTY)) {
String[] labels = Stream.concat(Stream.of(RESOURCE_LABEL), values.stream()
.map(String.class::cast)
.map(label -> prefixer.shorten(label, GraphNamingConvention.LABEL)))
.toArray(String[]::new);
visitor.labels(labels);
} else {
//TODO: Have values be Neo4J storable.values to avoid multiple array copies.
Object value = values.get(0);
if (value instanceof String) {
visitor.property(prefixer.shorten(key, GraphNamingConvention.PROPERTY_KEY), values.toArray(new String[values.size()]));
} else if (value instanceof Boolean) {
visitor.property(prefixer.shorten(key, GraphNamingConvention.PROPERTY_KEY), values.toArray(new Boolean[values.size()]));
} else if (value instanceof Integer) {
visitor.property(prefixer.shorten(key, GraphNamingConvention.PROPERTY_KEY), values.toArray(new Integer[values.size()]));
} else if (value instanceof Long) {
visitor.property(prefixer.shorten(key, GraphNamingConvention.PROPERTY_KEY), values.toArray(new Long[values.size()]));
} else if (value instanceof Double) {
visitor.property(prefixer.shorten(key, GraphNamingConvention.PROPERTY_KEY), values.toArray(new Double[values.size()]));
} else if (value instanceof Byte) {
visitor.property(prefixer.shorten(key, GraphNamingConvention.PROPERTY_KEY), values.toArray(new Byte[values.size()]));
} else if (value instanceof Float) {
visitor.property(prefixer.shorten(key, GraphNamingConvention.PROPERTY_KEY), values.toArray(new Float[values.size()]));
} else if (value instanceof Character) {
visitor.property(prefixer.shorten(key, GraphNamingConvention.PROPERTY_KEY), values.toArray(new Character[values.size()]));
} else if (value instanceof Short) {
visitor.property(prefixer.shorten(key, GraphNamingConvention.PROPERTY_KEY), values.toArray(new Short[values.size()]));
} else {
throw new RuntimeException("Property Map Entry with un-serializable type, Sample entry: " + value.toString());
}
}
}
}
visitor.endOfEntity();
return true;
} else {
return false;
}
}
@Override
public void close() throws IOException {
// not relevant
}
@Override
public void setStream(Stream<NodeWritable> newChunk) {
nodeWritableStream = newChunk.iterator();
}
}
The final piece of the puzzle is the parser. Our InputChunkIterator expects both a chunk and a parser with which to populate the chunk. As we were using a Windows machine for this test we’re explicitly loading the Windows DLLs however it is trivial to modify this to work for Linux.
public class NodeFileParser implements ChunkFileParser<NodeWritable> {
private static final Logger LOG = LoggerFactory.getLogger(NodeFileParser.class);
private static final Configuration HDFS_CONF = new Configuration(true);
private static final int AVERAGE_NODES_PER_FILE = 800000;
static {
System.load("C:/ProgramData/Hadoop/bin/hadoop.dll");
System.load("C:/ProgramData/Hadoop/bin/hdfs.dll");
System.setProperty("HADOOP_COMMON_LIB_NATIVE_DIR", "C:/ProgramData/Hadoop/bin");
System.setProperty("HADOOP_OPTS", "-Djava.library.path=C:/ProgramData/Hadoop/bin");
}
/*
* Method to read from a Sequence File of (String, NodeWritable)
*/
@Override
public Stream<NodeWritable> readFile(File file) throws IOException {
SequenceFile.Reader sfr = new SequenceFile.Reader(HDFS_CONF, SequenceFile.Reader.file(new Path(file.toURI())));
AtomicBoolean closed = new AtomicBoolean(false);
AtomicLong done = new AtomicLong(0);
return StreamSupport.stream(new Spliterators.AbstractSpliterator<NodeWritable>(AVERAGE_NODES_PER_FILE, Spliterator.DISTINCT | Spliterator.IMMUTABLE | Spliterator.NONNULL) {
@Override
public boolean tryAdvance(Consumer<? super NodeWritable> action) {
if (action == null) {
throw new NullPointerException("Cannot act on nothing");
}
if (!closed.get()) {
NodeWritable nodeWritable = new NodeWritable();
Text key = new Text();
boolean hadNext;
long doneNow = done.getAndIncrement();
if (doneNow % 100000 == 0) {
LOG.info("Read {}", doneNow);
}
try {
hadNext = sfr.next(key, nodeWritable);
} catch (IOException e) {
LOG.info("Failed to read next line in {}", file.toString());
throw new RuntimeException("Failed to read next line in file", e);
}
if (!hadNext) {
try {
LOG.info("Completed file after {}", done.get());
closed.compareAndSet(false, true);
sfr.close();
} catch (IOException e) {
e.printStackTrace();
}
return false;
}
action.accept(nodeWritable);
return true;
}
return false;
}
}, false);
}
}
To avoid the code listings getting out of hand I have not provided the interfaces nor the relationship section, however this is trivial as it is mostly cut and paste. This bulk interface also provides support for pre-prepping an index and much more besides.
Potential Gains
Even though there are further possible improvements, the amount of time taken by the Node phase of the loader is actually not particularly significant. In my latest run loading all nodes took a mere 5 minutes and 32 seconds!
Limitations and further work
The biggest limitation is this must be done entirely offline, however this isn’t a huge issue. We can resolve it trivially by simply having hot and cold instances. Much like a lambda architecture, we can re-compute a new store on the cold instance while still applying writes from streaming sources on the hot. When the cold is ready we replay all the writes it has missed and once it is in-sync with the hot instance simply switch the load balancer and allow all queries to switch to the other instance. This is a little more complex with a clustered setup but is still fine. You simply switch half of the read-replicas over to the new instance then flip the load balancer and reattach the other half.
Can it ever scale horizontally?
As discussed earlier, Neo4J Enterprise Edition includes Fabric which allows splitting the data across many shards. They have recently demonstrated the ability of Fabric to support a graph with over one trillion relationships. They utilise a similar set of principles as explored in this blog but as they are using Fabric they can shard the loading process across many nodes and don’t need to be so careful with the loading format.
We avoided this option as we did not want to expose the extra complexity of the sharding to the users for this specific use-case, nevertheless this proves that the system can scale past single machines if needed and is well worth reading about for anyone trying to scale the Neo4J write path.
It is important to note that without Enterprise edition, Neo4J does have limits on the size of graph it can store, and with community edition the limits are more severe. The operations manual includes the limits of the various store types. In community edition you cannot use the high_limit store and as such cannot store more than 34 billion nodes and relations or 68 billion properties. That said, most users will likely never exceed that limit and loading anything approaching that amount of data will benefit significantly from the approach detailed in this article.
Other improvements
Throughout this article I have hinted at other possible improvements. There will be a follow-up article which will explore a number of these as well as some more esoteric options including using Javascript on GraalVM to interact with this API and create a working Graph.
